For too long, cutting-edge medical AI technology has been confined behind costly commercial licenses and opaque "black box" systems. This has discouraged many research organizations, small and medium-sized development teams, and frontline physicians, slowing the process of technological innovation and equitable adoption. Now, a program called OpenMed of a new program is trying to break the logjam. The project is in Hugging Face community released over 380 advanced medical and clinical text-based named entity recognition (NER) models and announced that based on the Apache 2.0 The license is free and open in perpetuity.
The significance of this move lies not only in providing a free alternative, but also in the fact that the models it released outperformed even mainstream commercial closed-source solutions in multiple benchmarks. This signals that the technical barriers in healthcare AI are being dismantled by the power of open source.
Industry Dilemmas and Open Source Solutions
The healthcare AI field faces several core barriers to growth:
- High license fees: Top commercial AI tools are expensive to license, excluding academic institutions and startups with limited budgets.
- Technical opacity: Commercial tools typically do not disclose their model architecture, training data, and workings, making it difficult for users to assess their reliability and potential bias.
- Slow technology iteration: Some of the paid models have failed to keep up with the latest advances in AI technology, and their performance has gradually lagged behind cutting-edge research in academia.
- limited application: Quality AI capabilities are concentrated in the hands of a few large enterprises, limiting technology inclusion.
OpenMed The project responds directly to these challenges. It offers more than 380 free NER models that focus on recognizing key entities in medical texts such as drug names, diseases, genes, anatomical structures, etc. These models possess the following salient features:
- ✅ Totally free: Based on
Apache 2.0License that allows free use, modification and distribution. - ✅ ready-to-use: Designed for real-world scenarios, it can be deployed without a lot of extra work.
- ✅ Size FlexibilityThe number of model parameters ranges from 109M to 568M to accommodate different hardware requirements.
- ✅ proven: Rigorous performance testing on over 13 standardized datasets in the medical domain.
- ✅ ecologically compatible:: In conjunction with the
Hugging Face和PyTorchSeamless integration with mainstream frameworks such as
OpenMed Toolkit Details
OpenMed The model libraries have been carefully fine-tuned and tested in the Gellus F1 scores on datasets such as 0.998, demonstrating its superior performance.
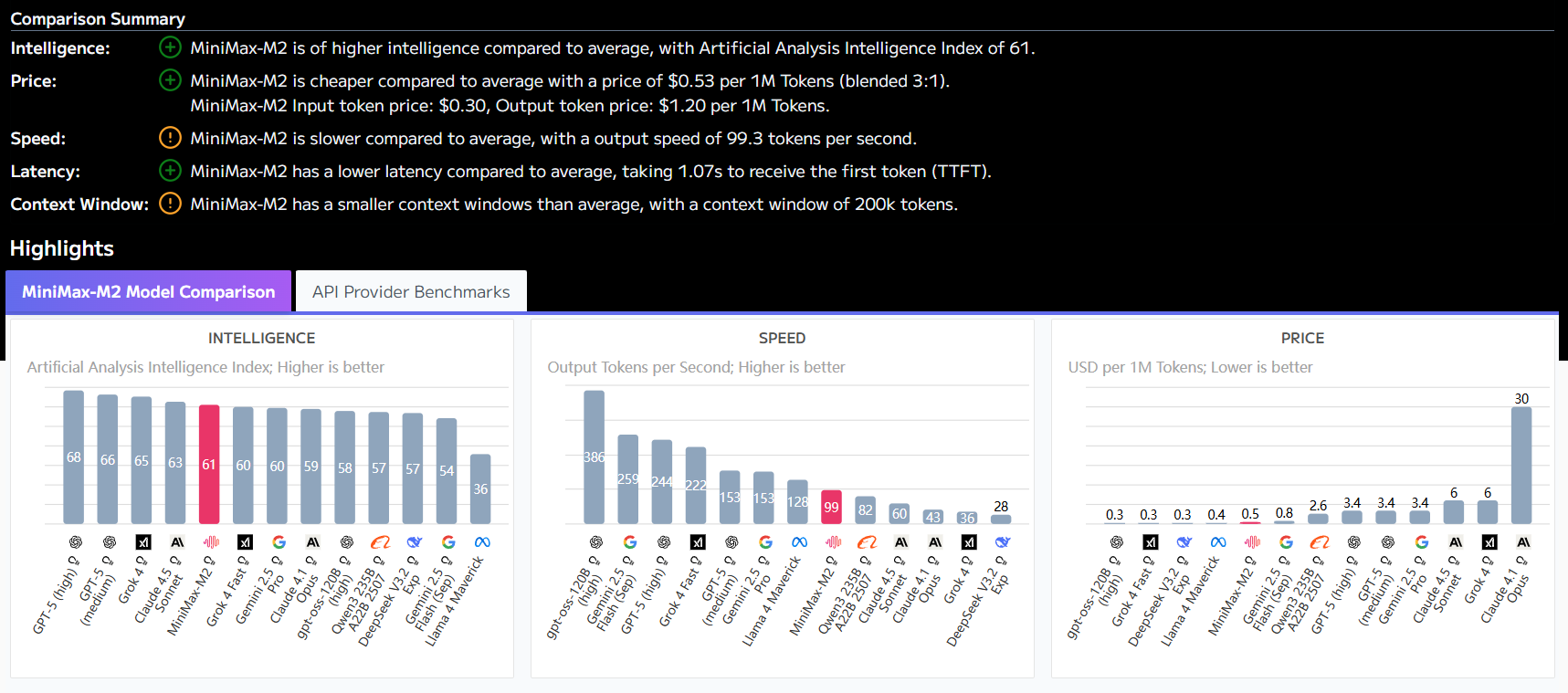
🔬 Performance comparison: open source OpenMed vs. closed-source business models
In order to visualize its competitiveness, theOpenMed published performance comparisons with current state-of-the-art closed-source business models. The data shows thatOpenMed Not only is it comparable to commercial models on several datasets, but it also achieves significant outperformance in certain scenarios.
| data set | OpenMed Best F1 Score (%) | Closed source SOTA F1 score (%)† | Gap (OpenMed - SOTA) | current closed-source leader |
|---|---|---|---|---|
| BC4CHEMD | 95.40 | 94.39 | +1.01 | Spark NLP BertForTokenClassification |
| BC5CDR-Chem | 96.10 | 94.88 | +1.22 | Spark NLP BertForTokenClassification |
| BC5CDR-Disease | 91.20 | 88.5 | +2.70 | BioMegatron |
| NCBI-Disease | 91.10 | 89.71 | +1.39 | BioBERT |
| JNLPBA | 81.90 | 82.00 | –0.10 | KeBioLM (knowledge-enhanced LM) |
| Linnaeus | 96.50 | 92.70 | +3.80 | BERN2 toolkit |
| Species-800 | 86.40 | 82.59 | +3.81 | Spark NLP BertForTokenClassification |
| BC2GM | 90.10 | 88.75 | +1.35 | Spark NLP Bi-LSTM-CNN-Char |
| AnatEM | 90.60 | 91.65 | –1.05 | Spark NLP BertForTokenClassification |
| BioNLP 2013 CG | 89.90 | 87.83 | +2.07 | Spark NLP BertForTokenClassification |
| Gellus | 99.80 | 63.40 | +36.40 | ConNER |
| CLL | 95.70 | 85.98 | — | (No SOTA issued) |
| FSU | 96.10 | — | — | (No SOTA issued) |
† Closed-source scores are derived from the highest published peer-reviewed or ranked results in the literature (typically commercial models such as Spark NLP, NEEDLE, BERN2, etc.).
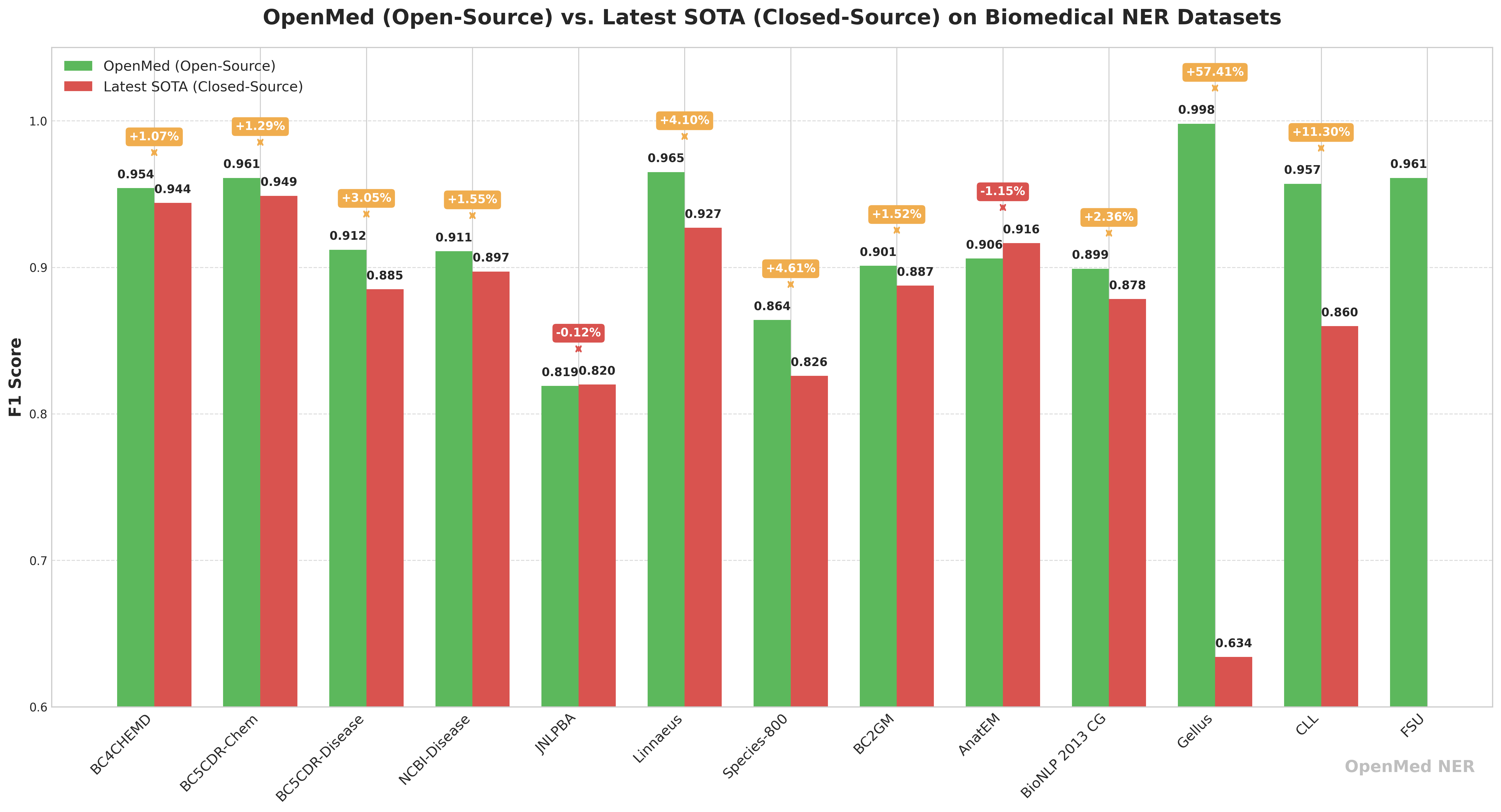
Of particular concern is the fact that the Gellus on the dataset.OpenMed The F1 score is 36.41 TP3T higher than the previous best model, which shows that open source models focused on optimization have great potential on specific tasks.
🔬 By field of application
The following table maps the datasets to their corresponding healthcare domains and recommends the appropriate models based on the combined performance of the datasets in each domain.
| realm | Included data sets | Number of available models | Range of parameters | recommendation model |
|---|---|---|---|---|
| pharmacology | bc5cdr_chem, bc4chemd, fsu |
90 | 109M – 568M | OpenMed-NER-PharmaDetect-SuperClinical-434M |
| Disease/Pathology | bc5cdr_disease, ncbi_disease |
60 | 109M – 434M | OpenMed-NER-PathologyDetect-PubMed-v2-109M |
| genomics | jnlpba, bc2gm, species800, linnaeus, gellus |
150 | 335M – 568M | OpenMed-NER-GenomicDetect-SnowMed-568M |
| anatomical | anatomy |
30 | 560M | OpenMed-NER-AnatomyDetect-ElectraMed-560M |
| study of tumors | bionlp2013_cg |
30 | 355M | OpenMed-NER-OncologyDetect-SuperMedical-355M |
| clinical record | cll |
30 | 560M | OpenMed-NER-BloodCancerDetect-ElectraMed-560M |
⚡️ Select by model size
| magnitude | quantity of participants | Optimal scenarios |
|---|---|---|
| compact | 109M | Rapid prototyping and low resource environment |
| mega | 335M – 355M | Balanced choice of accuracy and performance |
| mega | 434M | All-rounder with excellent performance |
| giant | 560M – 568M | A mission for extreme precision |
📊 The best model on each dataset
Below is a summary of the best performing models on each dataset and their F1 scores and sizes.
| data set | best model | F1 Score | Model size (parameter) |
|---|---|---|---|
bc5cdr_chem |
OpenMed-NER-PharmaDetect-SuperClinical-434M |
0.961 | 434M |
bionlp2013_cg |
OpenMed-NER-OncologyDetect-SuperMedical-355M |
0.899 | 355M |
bc4chemd |
OpenMed-NER-ChemicalDetect-PubMed-335M |
0.954 | 335M |
linnaeus |
OpenMed-NER-SpeciesDetect-PubMed-335M |
0.965 | 335M |
jnlpba |
OpenMed-NER-DNADetect-SuperClinical-434M |
0.819 | 434M |
bc5cdr_disease |
OpenMed-NER-DiseaseDetect-SuperClinical-434M |
0.912 | 434M |
fsu |
OpenMed-NER-ProteinDetect-SnowMed-568M |
0.961 | 568M |
ncbi_disease |
OpenMed-NER-PathologyDetect-PubMed-v2-109M |
0.911 | 109M |
bc2gm |
OpenMed-NER-GenomeDetect-SuperClinical-434M |
0.901 | 434M |
cll |
OpenMed-NER-BloodCancerDetect-ElectraMed-560M |
0.957 | 560M |
gellus |
OpenMed-NER-GenomicDetect-SnowMed-568M |
0.998 | 568M |
anatomy |
OpenMed-NER-AnatomyDetect-ElectraMed-560M |
0.906 | 560M |
species800 |
OpenMed-NER-OrganismDetect-BioMed-335M |
0.864 | 335M |
Getting Started Fast and Scaling Applications
leverage Hugging Face Transformers libraries, integrating OpenMed The modeling process is very simple and can be invoked with just three lines of code.
from transformers import pipeline
ner_pipeline = pipeline("token-classification", model="OpenMed/OpenMed-NER-PharmaDetect-SuperClinical-434M", aggregation_strategy="simple")
text = "Patient prescribed 10mg aspirin for hypertension."
entities = ner_pipeline(text)
print(entities)
# 输出: [{'entity_group': 'CHEMICAL', 'score': 0.99..., 'word': 'aspirin', 'start': 28, 'end': 35}]
For scenarios where large-scale datasets need to be processed, the project also provides efficient batch processing solutions.
from transformers.pipelines.pt_utils import KeyDataset
from datasets import Dataset, load_dataset
import pandas as pd
# 加载公开的医疗数据集(使用一个子集进行测试)
medical_dataset = load_dataset("BI55/MedText", split="train[:100]")
data = pd.DataFrame({"text": medical_dataset["Completion"]})
dataset = Dataset.from_pandas(data)
# 使用适合您硬件的批处理大小
batch_size = 16 # 根据您的 GPU 显存进行调整
results = []
ner_pipeline = pipeline("token-classification", model="OpenMed/OpenMed-NER-PharmaDetect-SuperClinical-434M", device=0) # 使用GPU
for out in ner_pipeline(KeyDataset(dataset, "text"), batch_size=batch_size):
results.extend(out)
print(f"批处理完成 {len(results)} 条文本")
NER The Real World Value of Technology Unlocked
Named Entity Recognition (NER) technology automatically extracts and categorizes key information from unstructured text. In healthcare, this technology is a catalyst for activating the value of massive amounts of data in clinical notes, patient records and scientific literature.
- 🔒 Patient privacy protection (data de-identification):
NERPersonal health information (PHI) such as name, address, etc. can be automatically recognized and removed from medical records. This is important in protecting patient privacy, complying withHIPAAand other laws and regulations, while providing a compliant and secure source of data for medical research that is far more efficient and accurate than manual processing. - 🔗 Medical Knowledge Graph Construction (Entity Relationship Extraction): After identifying entities such as drugs, diseases, etc., further techniques can analyze the relationships between them (e.g., "Drug A causes Side Effect B"). This helps to build a medical knowledge graph that supports clinical decision-making, accelerates drug development, and ultimately enables personalized treatment.
- 💡 Optimizing Healthcare Costs and Management (HCC Codes): Hierarchical Condition Category (HCC) coding is a key process used by healthcare payers, such as Medicare, to project costs and set reimbursement rates.
NERDiagnostic information can be automatically extracted from medical records to aid in coding, ensuring that providers are fairly compensated for treating complex cases while helping to identify high-risk patients for proactive intervention.
By promoting the automation of these critical tasks, theNER Technology is transforming dormant medical texts into actionable solutions that enhance data security, accelerate research, improve patient prognosis and reduce operational costs.OpenMed The emergence of a new system will undoubtedly accelerate the process considerably.