GBC MedAI is an open source intelligent medical assistant system. It uses FastAPI as the backend framework and Vue 3 as the frontend technology stack. The core capability of this system lies in the ability to integrate multiple AI big language models at the same time, such as DeepSeek, locally deployed Ollama and other models compatible with the OpenAI interface. It also integrates a variety of search engine services to provide users with intelligent dialog and search functions that incorporate real-time information. With a modular architecture and an advanced and complete technology stack, the project aims to provide an AI assistant solution that can be rapidly deployed and secondarily developed for the medical field or other professional scenarios.
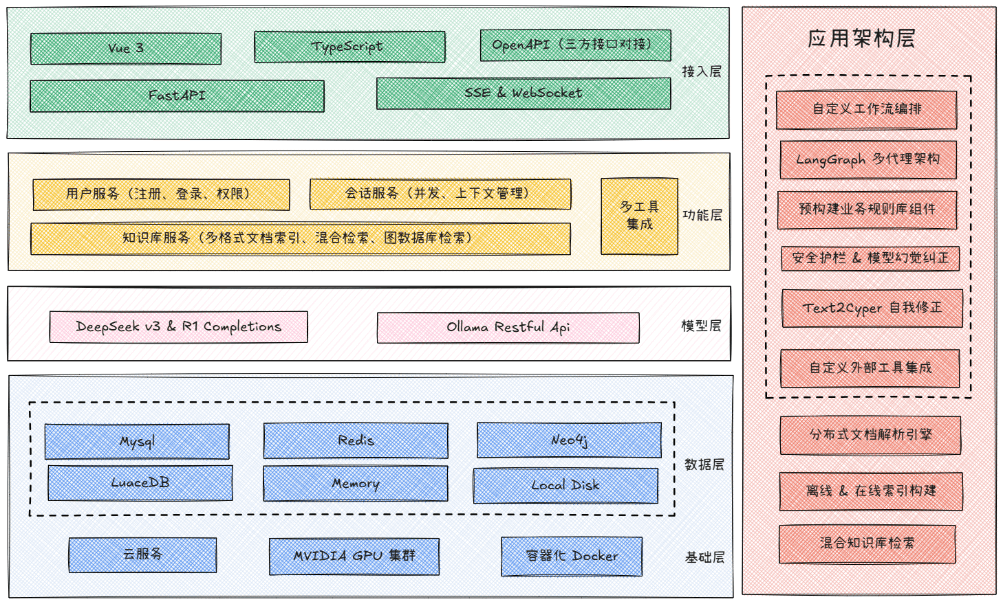
Function List
- AI capabilities: It supports access to DeepSeek, Ollama local models, and a variety of AI models compatible with OpenAI. The system has the ability to handle complex reasoning tasks and visual understanding, and can analyze uploaded images.
- Intelligent Search: Integration of Bucha AI, Baidu AI Search and SerpAPI, enabling automatic selection of the most appropriate search engine based on the question and access to the latest medical information and research progress.
- dialog system:: Provides smooth streaming response and contextual memory features. The system has built-in complete user authentication and multi-session v-session management mechanism to save history.
- Modernization Front End: Built with Vue 3 and TypeScript, it provides a responsive layout and supports both desktop and mobile devices. The interface is designed in a beautiful secondary anime style and includes rich interactive animations.
- High-performance back-end: Developed on the FastAPI asynchronous framework, it ensures efficient request processing capabilities.
- Data and Caching: Using Redis as a semantic cache can effectively improve the response performance of repetitive queries and reduce the cost of AI model calls.
- Advanced AI framework:: Applied LangGraph and technologies such as GraphRag for building more complex AI processes and knowledge graph applications.
Using Help
This section details how to install, configure and successfully run the GBC MedAI system. Please follow the steps below to complete your deployment.
Environmental requirements
Before you begin, make sure your system environment meets the following conditions:
- Python: 3.8 or higher
- Node.js: 16 or higher (required only if front-end development is required)
- comprehensive database: MySQL 8.0 or higher, Redis 6.0 or higher
- Knowledge Graph (optional): Neo4j 4.0 or higher
Prefile Download
The project relies on some preconfigured files to run, please download and unzip them into the specified directory from the following address first:
- Front-end documentation: Link from Baidu.com
https://pan.baidu.com/s/1KTROmn78XhhcuYBOfk0iUQ?pwd=wi8k(Extract code: wi8k) Download the front-end package file and extract it to thellm_backend/static/Catalog. - GraphRag files: Link from Baidu.com
https://pan.baidu.com/s/1dg6YxN_a4wZCXrmHBPGj_Q?pwd=hppu(Extract code: hppu) Download the GraphRag related files and place them in the project root directory.
Installation and configuration steps
1. Cloning of project codes
First, use the git command to clone the project code from GitHub to your local computer.
git clone https://github.com/zhanlangerba/gbc-madai.git
cd gbc-madai
2. Back-end service setup
Go to the backend code directory and use the pip Install all Python dependencies.
cd llm_backend
pip install -r requirements.txt
3. Configuring environment variables
The configuration file is key to the operation of the system. You will need to copy the template file and fill in your personal configuration information.
# 复制环境变量模板文件
cp .env.example .env
# 使用文本编辑器(如 nano 或 vim)打开并编辑 .env 文件
nano .env
在 .env document, you will need to fill in the following information:
DEEPSEEK_API_KEY: Your DeepSeek API key.BOCHA_AI_API_KEY/BAIDU_AI_SEARCH_API_KEY/SERPAPI_KEY: Fill in the corresponding API key according to the search engine you have chosen.DB_HOST,DB_USER,DB_PASSWORD,DB_NAME: Your MySQL database connection information.REDIS_HOST,REDIS_PORT: Your Redis service connection information.NEO4J_URL,NEO4J_USERNAME,NEO4J_PASSWORD: Your Neo4j database connection information (if used).
4. Initialization of the database
Before starting the service, you need to create the database required for the project in MySQL.
# 使用您的 MySQL root 用户登录并执行以下命令
mysql -u root -p -e "CREATE DATABASE assist_gen CHARACTER SET utf8mb4 COLLATE utf8mb4_unicode_ci;"
5. Starting system services
After completing all the above steps, run run.py script to start the entire system.
# 启动后端服务
python run.py
After the service has been successfully started, you can access the following address via your browser:
- front end access address:
http://localhost:8000/ - API Documentation Address (Swagger UI):
http://localhost:8000/docs
Deployment method
In addition to running the source code directly, the project supports the more stable and convenient Docker deployment method.
Deploying with Docker (recommended)
Ensure that you have installed Docker. execute the following command in the root directory of your project:
# 1. 构建 Docker 镜像,将其命名为 gbc-madai
docker build -t gbc-madai .
# 2. 以后台模式运行容器,并将本地的 .env 文件作为环境变量传入
docker run -d -p 8000:8000 --env-file .env gbc-madai
Independent front-end development
If you need to modify and develop the front-end interface, you can go to the gbc_madai_web directory to run the front-end development service independently. "`shell
Go to the front-end code directory
cd ../gbc_madai_web
Installing front-end dependencies
npm install
Start the front-end development server
npm run dev
Front-end development services will be available in http://localhost:3000 starts and will automatically proxy API requests to the http://localhost:8000。
application scenario
- Intelligent Healthcare Consulting
As a virtual physician's assistant, it can be used 24/7 online to answer patients' questions about common illnesses, medication advice and healthy living, and provide initial medical guidance. - Clinical decision support
Assisting doctors to quickly retrieve and analyze the latest medical literature, clinical trial reports and treatment guidelines, providing information support for the development of more accurate diagnosis and treatment plans. - Medical Information Retrieval Platform
It can be used as an intelligent knowledge base within a hospital or research organization to help staff quickly find internal rules and regulations, operating procedures, case files and research materials. - Other specialized field applications
The modular design of the system allows it to be easily extended to other professional fields such as e-commerce, finance, law, etc., to be used as intelligent customer service or expert assistant.
QA
- Is this project free and open source?
Yes, the project is completely open source based on the MIT license and you are free to use, modify and distribute it in your personal or commercial projects. - What technical background is required to deploy this system?
You will need a basic understanding of the Python language and the FastAPI framework, as well as knowledge of MySQL and Redis database installation and basic operations. If you choose to deploy with Docker, you will also need to familiarize yourself with common Docker commands. - Can I access my own AI models?
Possible. The system architecture supports access to any model that is compatible with the OpenAI API standard. All you need to do is add the.envModify the model's API address in the configuration file (OLLAMA_BASE_URL(or similar variables) and a key is sufficient. The system itself has native support for DeepSeek and Ollama. - Can the front-end interface be replaced?
The system backend provides a complete set of REST APIs. The system back-end provides a complete set of REST APIs. you can skip the front-end that comes with your project altogether and follow the API documentation (http://localhost:8000/docs) Develop your own client applications, such as desktop programs, mobile apps, or applets.