Recently, OpenAI and biotech startup Retro Biosciences announced the results of a collaboration that demonstrates the enormous potential of artificial intelligence in the life sciences. Using an AI model, GPT-4b micro, designed specifically for protein engineering, the team succeeded in improving the expression efficiency of key markers for induced stem cell reprogramming by more than 50-fold.
The core of this research focuses on the Yamanaka factors, a group of proteins that won the Nobel Prize for their pioneering role in cell reprogramming. These proteins can turn differentiated adult cells, such as skin cells, into "induced pluripotent stem cells" (iPSCs) with the potential to develop into virtually any tissue. This ability opens up new avenues for treating blindness, diabetes, infertility, and even solving organ shortages.
However, the application of traditional Yamanaka factors is extremely inefficient, with typically less than 0.1% of cells being successfully transformed, and the entire process taking more than three weeks. The efficiency is further reduced especially when dealing with cells from older or diseased donors. This time, the AI-redesigned protein variant not only significantly improved efficiency, but also demonstrated enhanced DNA damage repair capabilities, which means it has greater potential for cell rejuvenation.
This initial discovery, made in 2025, has now been validated by replicated experiments in a variety of donors, cell types, and delivery methods, confirming the complete pluripotency and genomic stability of the resulting iPSC cell lines.
Experimental GPT models tailored for protein engineering
To validate that AI can accelerate life science research, OpenAI has built a custom model called GPT-4b micro. The model is a miniature version of GPT-4o, specially trained to give it deep biological knowledge, especially in the areas of protein engineering, control and flexibility.
Unlike most protein language models, the training data for GPT-4b micro contains not only protein sequences, but also incorporates biological text and tagged 3D structural data. The training data is particularly enriched with contextual information such as textual descriptions of proteins, co-evolved homologous sequences, and known interacting proteomes. This approach allows the model to generate sequences based on cues from specific attributes, and to handle structured proteins and "intrinsically disordered" proteins equally well. Yamanaka factors are the latter, and their activity depends on a large number of transient interactions with multiple binding partners rather than a fixed single stable structure.
In this way, the effective context length of the model far exceeds the limit of independent sequences, and up to 64,000 can be processed in the inference process token cues, which is unprecedented in protein sequence modeling.
Artificial Intelligence Assisted Retrofit SOX2 and KLF4
Yamanaka factor consists of four proteins: OCT4, SOX2, KLF4 and MYC (OSKM for short). Optimizing them directly by modifying the protein sequences is a difficult task. In the case of SOX2 (containing 317 amino acids) and KLF4 (containing 513 amino acids), for example, the number of possible variants is up to 10 to the 1000th power.
The traditional "directed evolution" approach, in which only a few amino acid residues are changed at a time, only explores a tiny fraction of the possibilities. AI, by contrast, can explore a much wider design space. The Retro Biosciences team first set up a wet-lab screening platform and then used the GPT-4b micro to generate a series of candidate sequences called "RetroSOX".
The results were surprising: in the screen, model-suggested sequences exceeding 30% outperformed wild-type SOX2 in the expression of key pluripotency markers, even though their amino acid sequences differed by more than 100 on average. In contrast, in conventional screens, hits were typically below 10% .
Next, the team targeted KLF4. The model generated 14 "RetroKLF" variants that outperformed the best combination in the RetroSOX screen, with a hit rate close to 50%.
The effect was most dramatic when the top RetroSOX and RetroKLF variants were combined. In three independent experiments, both early and late pluripotency markers in fibroblasts increased dramatically, and the late markers appeared several days earlier than with the wild-type OSKM mixture. Further tests, such as alkaline phosphatase (AP) staining, also confirmed that these cell colonies not only expressed late phase markers, but also exhibited robust AP activity, a strong indicator of pluripotency.
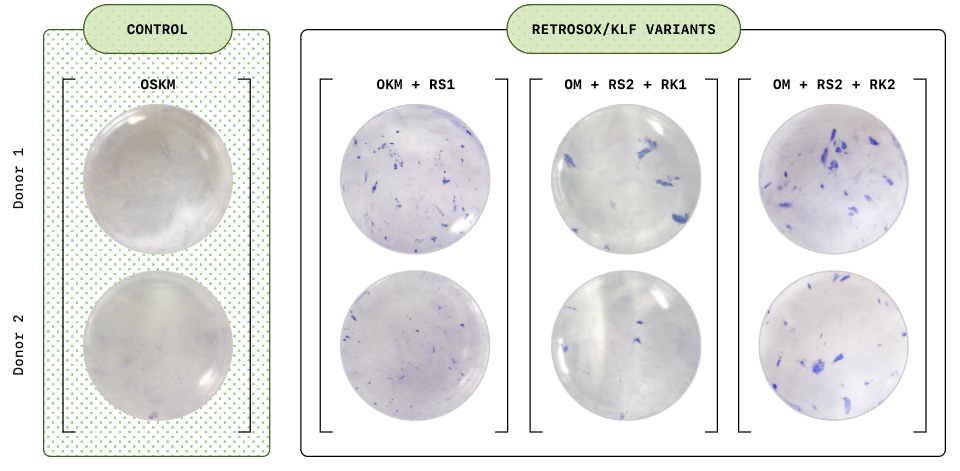
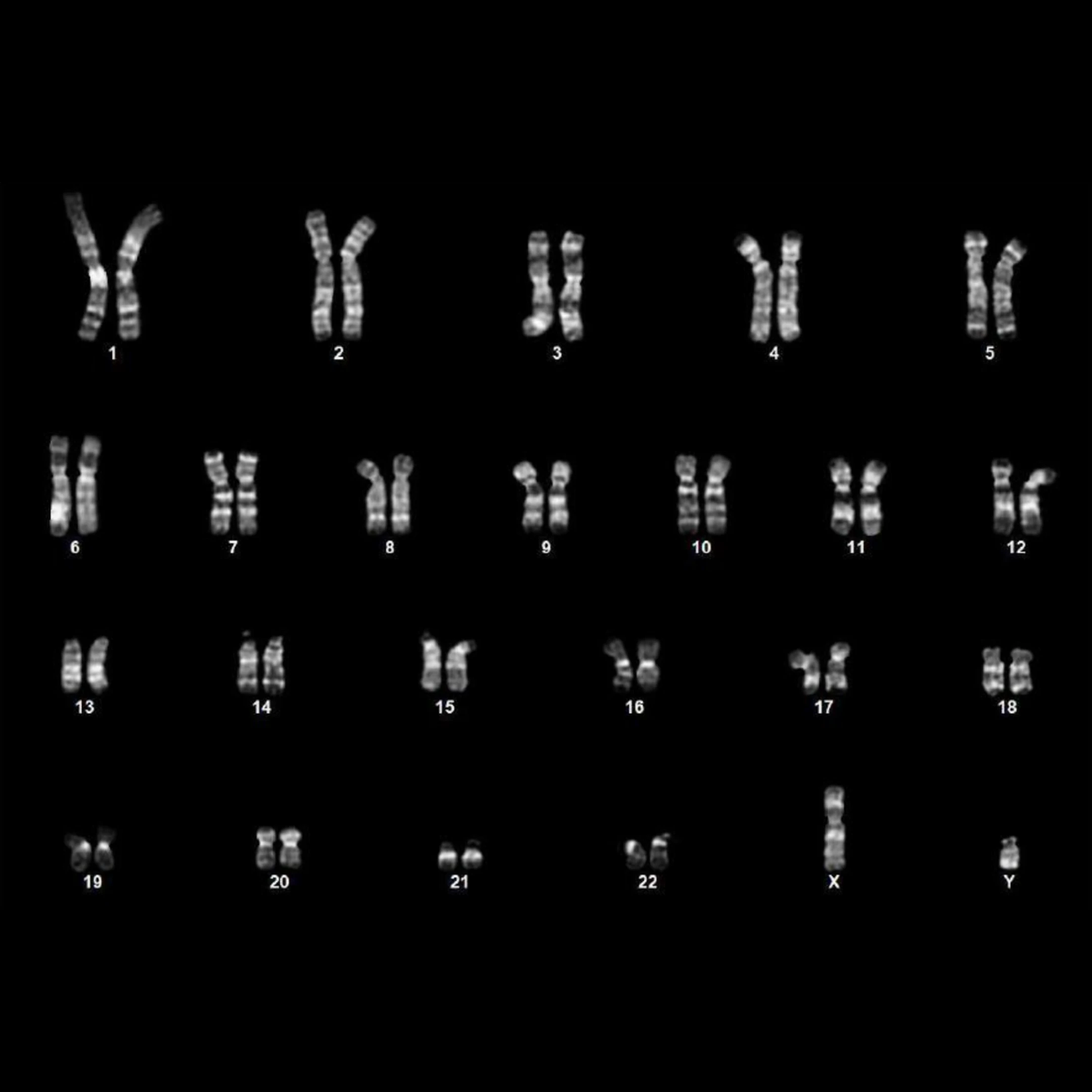
To explore the clinical potential, the team also tested a different delivery method (mRNA alternative to viral vectors) and another cell type - mesenchymal stromal cells (MSCs) from three middle-aged donors over the age of 50. Within only 7 days, more than 30% cells began to express key pluripotency markers; by day 12, more than 85% cells activated endogenous stem cell markers including OCT4, NANOG. Karyotyping of these cells showed normal chromosome structure, confirming their genomic stability and suitability for cell therapy.
Enhanced DNA damage repair
In addition to improving reprogramming efficiency, the researchers explored the potential of these engineered variants for cellular rejuvenation, particularly the ability to repair DNA damage, one of the classic hallmarks of cellular senescence.
In DNA damage analysis, after treatment with genotoxic chemicals, cells expressing the RetroSOX/KLF mixture displayed significantly lower DNA double-strand break markers (γ-H2AX signaling) than cells using standard OSKM or controls. This suggests that the AI-designed protein variants can repair DNA damage more efficiently, providing a new possible pathway for delaying cellular senescence.

future outlook
This work clearly demonstrates how quickly a domain-specific AI model can achieve breakthroughs on focused scientific problems. When researchers combine deep domain insights with language modeling tools, problems that once took years to solve may now progress in days.
Of course, this research is still in the early stages, and safety and long-term effects still need to be carefully evaluated before moving from the lab to clinical applications. But it undoubtedly opens a new door for the application of AI in the biomedical field, heralding the arrival of a new era of AI-driven personalized medicine and regenerative medicine.